Associate Research Fellow Li Dong homepage
Key Laboratory for NeuroInformation, Ministry of Education, China
♦ Master Qiankun Xie provides the basic NIT v1.0 code.
♦ Thanks the SPM, EEGLAB, MRIcro and DPARSF software.
♦ Cite the toolbox
###################<Please cite the toolbox as>###################
Li Dong*, Cheng Luo*, Xiaobo Liu, Sisi Jiang, Fali Li, Hongshuo Feng, Jianfu Li, Diankun Gong and Dezhong Yao*. Neuroscience Information Toolbox: an open source toolbox for EEG-fMRI multimodal fusion analysis. Frontiers in Neuroinformatics, 2018, 12:56. doi: 10.3389/fninf.2018.00056.
♦ Original papers
Dong, L., et al., 2014. Simultaneous EEG-fMRI: Trial level spatio-temporal fusion for hierarchically reliable information discovery. Neuroimage. 99, 28-41.
Dong, L., et al., 2015a. Spatio-temporal Consistency of Local Neural Activities: A New Imaging Measure for functional Magnetic Resonance Data. Journal of Magnetic Resonance Imaging. 42, 729-36.
Dong, L., et al., 2015b. Local Multimodal Serial Analysis for Fusing EEG-fMRI: A New Method to Study Familial Cortical Myoclonic Tremor and Epilepsy. Autonomous Mental Development, IEEE Transactions on. PP, 1-1.
Dong, L., et al., 2017. MATLAB Toolboxes for Reference Electrode Standardization Technique (REST) of Scalp EEG. Frontiers in Neuroscience. 11.
Lei, X., et al., 2010. A parallel framework for simultaneous EEG/fMRI analysis: methodology and simulation. Neuroimage. 52, 1123-34.
Lei, X., et al., 2011. fMRI functional networks for EEG source imaging. Hum Brain Mapp. 32, 1141-60.
Strobel, A., et al., 2008. Novelty and target processing during an auditory novelty oddball: a simultaneous event-related potential and functional magnetic resonance imaging study. Neuroimage. 40, 869-83.
Tomasi, D., Volkow, N.D., 2010. Functional connectivity density mapping. Proc Natl Acad Sci U S A. 107, 9885-90.
♦ Related papers
I. Copyright ©
All copyright of the NIT software reserved by the Key Laboratory for NeuroInformation of Ministry of Education, School of Life Science and Technology, University of Electronic Science and Technology of China. This software is for non-commercial use only. It is freeware but not in the public domain.
II. NIT Toolbox Version Release
The NIT manual is contained in the toolbox
III. Example data
The fMRI data during 510 s resting state (two healthy adults, TR = 2 s) are provided as an example to show the NIT function. The whole brain mask is also provided.
Simultaneous EEG-fMRI recording data during an auditory novelty oddball (target processing) are provided as an example. Here, P300 was first extracted from preprocessed EEG data using temporal independent component analysis (ICA), and then NESOI was used to reconstruct the EEG source while using networks (fMRI spatial ICA components) as the covariance priors. Further details regarding the task were observed in a previous article (Dong et al., 2014; Strobel et al., 2008).
EEG_informed_fMRI_example_data
Simultaneous EEG and fMRI (TR = 2s) recording data during resting state in familial cortical myoclonic tremor and epilepsy (FCMTE) patients are provided as an example. The onset times of epileptic discharges which were independently identified by two experienced neurologists with the best agreement are provided instead of original EEG data. More details can be seen in the paper (Dong et al., 2015b).
P300_EEG_informed_fMRI_example
Simultaneous EEG and fMRI (TR = 2s) recording data during P300 visual task in a healthy subject are provided as another EEG-informed fMRI analysis example. More details of this example can be seen in read me.docx in the zip file.
IV. Results
The results of examples are provided below:
EEG_informed_fMRI_example_results
V. Other sources
Achievements of "The Key Laboratory for NeuroInformation of Ministry of Education"
Decoding the brain functions and dysfunctions continues to be grand challenge of the 21st century, and functional neuroimaging has shown tremendous promise in better understanding the brain (Friston, 2009). As yet, noninvasive imaging technologies such as Magnetic Resonance Image (MRI), electroencephalogram (EEG) and magnetoencephalogram (MEG) have been widely used in neuroimaging research. Considering their respective strengths and weaknesses in temporal and spatial resolution (see Fig. 1), integrating multimodal information (especially EEG-fMRI) may provide high-resolution spatiotemporal neuroimaging that further our understanding of the brain functions and dysfunctions (Biessmann et al., 2011; Huster et al., 2012; Laufs, 2012). For the EEG-fMRI fusion, there are the three most influential approaches for EEG-fMRI integration (He et al., 2011; Huster et al., 2012; Rosa et al., 2010) are the following (see Fig. 2): (1) fMRI-informed EEG, in which spatial information from the fMRI is utilized to assist the inverse problem of electromagnetic source reconstruction; (2) EEG-informed fMRI, in which the fMRI benefits from extracted EEG feature in specific frequency or time; and (3) symmetric EEG-fMRI fusion, in which EEG and fMRI data are analyzed jointly through a common generative model or in a common data space.

Fig. 1: The emotion of fusion: complementary advantages of spatial and temporal resolution.
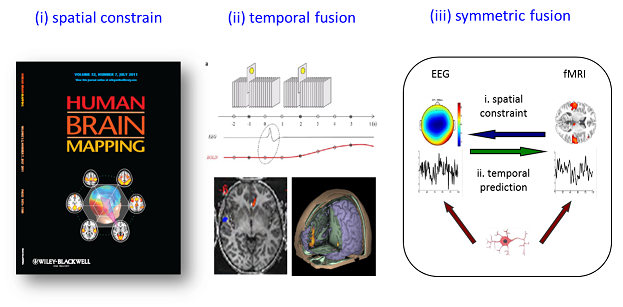
Fig. 2: Three most influential approaches for EEG-fMRI integration: (i) fMRI-informed EEG analysis; (ii) EEG-informed fMRI analysis; (iii) symmetric EEG-fMRI fusion.
♦ fMRI-informed EEG analysis
Spatial integration of EEG-fMRI typically utilizes the fMRI maps as a priori information to inform the locations of the EEG sources, such as fMRI-constrained dipole fitting and the fMRI-constrained/weighted source imaging. With the spatial constrains, the ill-posedness of the original inverse problem can be moderated. Considering brain networks represent the interactions between different brain areas, and understanding such networks may facilitate electroencephalography (EEG) source estimation. We proposed a new method, termed NEtwork-based SOurce Imaging (NESOI) (Lei et al., 2010; Lei et al., 2011), to reconstruct the EEG source using Parametric Empirical Bayesian (PEB). NESOI produces a good solution by combining the high temporal resolution of EEG and the high spatial resolution of fMRI. The NIT realizes the functions of NESOI.
♦ EEG-informed fMRI analysis
Temporal integration of EEG-fMRI typically utilizes the EEG dynamic features in the time or frequency domain, such as onset times of epileptic discharges, event related potentials, powers in the specific band etc., to inform the statistical mapping of fMRI. These quantities obtained from EEG are typically convolved with a canonical hemodynamic response function (HRF) and then regressed with BOLD signals on the voxel-level to identify the statistical fMRI activation/deactivation maps corresponding to the electromagnetic temporal signatures. A classic example is using onset times of epileptic discharges obtained from EEG to study the Blood Oxygenation Level Dependent (BOLD) changes related to the scalp EEG discharges, and providing useful information for epileptic mechanism research and clinical diagnosis (Gotman and Pittau, 2011; Murta et al., 2014).
However, there are also some problems for the analysis of simultaneous EEG-fMRI data in epilepsy: one is the variation of HRFs, and another is low signal-to-noise ratio in the data. Therefore, we proposed a new multimodal unsupervised method, termed local multimodal serial analysis (LMSA), which may compensate for these deficiencies in multimodal integration (Dong et al., 2015b).
♦ symmetric EEG-fMRI fusion
Symmetric EEG-fMRI fusion aims to benefit from both modalities while avoiding the bias of either method. One typical example of symmetric EEG-fMRI fusion is model-driven fusion such as dynamic causal models (DCM) (Friston et al., 2003; Kiebel et al., 2007). The merit of DCM is that it bases on a generative model which is neurophysiologically grounded; however, it always confronts with the complexity of the model and tedious calculations. In addition, data-driven symmetric fusion, which jointly assesses information gained from both modalities in a common data space (Moosmann et al., 2008) or feature space (Eichele et al., 2009) with joint independent component analysis (joint ICA) or parallel independent component analysis (parallel ICA), is also utilized.
22
NIT v1.3 (20180716):
♦Add two kinds of fMRI preprocessing pipelines for sequential acquisition:
Preprocess 6 (SPM12):realignment -> slice timing -> TPM normalise -> smooth
Preprocess 7 (SPM12):realignment -> slice timing -> T1SegNormalise -> smooth
♦Provide the head motion information based on mean framewise displacement (FD). It can be used to exclude subjects with large head motion.
♦In FOCA and FCD calculation, Power 12 head motion parameters will be regressed out.
♦Add FMRIB Plugin in EEG Toolbox for fMRI-related artifact removal.
♦Add an example of P300 EEG-informed fMRI analysis.
♦Revise codes and the manual
NIT (Neuroscience Information Toolbox) is a convenient toolkit for EEG-fMRI multimodal fusion, fMRI data preprocessing and analyzing. Using the NIT to analyze your data, you can do:
♦ Batch processing of the fMRI data analysis;
♦ Data preprocessing based on SPM12;
♦ Removing nuisance signals from fMRI data such as linear trend, head motion, white, CSF and global signals, and band-pass filtering;
♦ Calculate functional connectivity density (FCD) and dynamic FCD (sliding window);
♦ Calculate variability of dynamic timeseries/measures;
♦ Calculating FOur dimensional (spatio-temporal) Consistency of local neural Activities (FOCA);
♦ Parallel computing for nuisance signals removing, FCD and FOCA calculating;
♦ Extract features from cleaned EEG data, such as event onsets, power and ERP amplitudes;
♦ Marking interested events (e.g. epileptic discharges) in EEG data;
♦ EEG-fMRI multimodal fusion: 1) fMRI-informed EEG analysis (NEtwork-based SOurce Imaging, NESOI); 2) EEG-informed fMRI analysis (GLM and LMSA).
♦ Identify and classify original DICOM data;
♦ Viewing threshold of T-map.
Download the NIT toolbox and example data would be helpful for knowing more about how to use this software (The latest release is NIT_v1.3_20180716). Add NIT's directory to MATLAB's path and enter "nit" in the command window of MATLAB to enjoy it.
Neuroscience Information Toolbox

♦ Cite the toolbox
###################<Please cite the toolbox as>###################
Li Dong*, Cheng Luo*, Xiaobo Liu, Sisi Jiang, Fali Li, Hongshuo Feng, Jianfu Li, Diankun Gong and Dezhong Yao*. Neuroscience Information Toolbox: an open source toolbox for EEG-fMRI multimodal fusion analysis. Frontiers in Neuroinformatics, 2018, 12:56. doi: 10.3389/fninf.2018.00056.